Performant Clustering of Geo Coordinates w/ Custom Distance Functions
Recently I worked on a project triangulating geo-coordinates of signals based on registrations in a radio network. Part of the approach involved clustering observations into distinct geographic regions. The clustering algorithms used weren’t anything special (nearest-neighbor, k-means), however measuring the distance between points in geo-coordinates is not as simple as measuring the distance between points in cartesian coordinates.
Instead of the euclidean distance supported by most clustering algorithms, the ideal metric is Haversine Distance which measures the shortest path between two points traveling along the surface of a sphere.
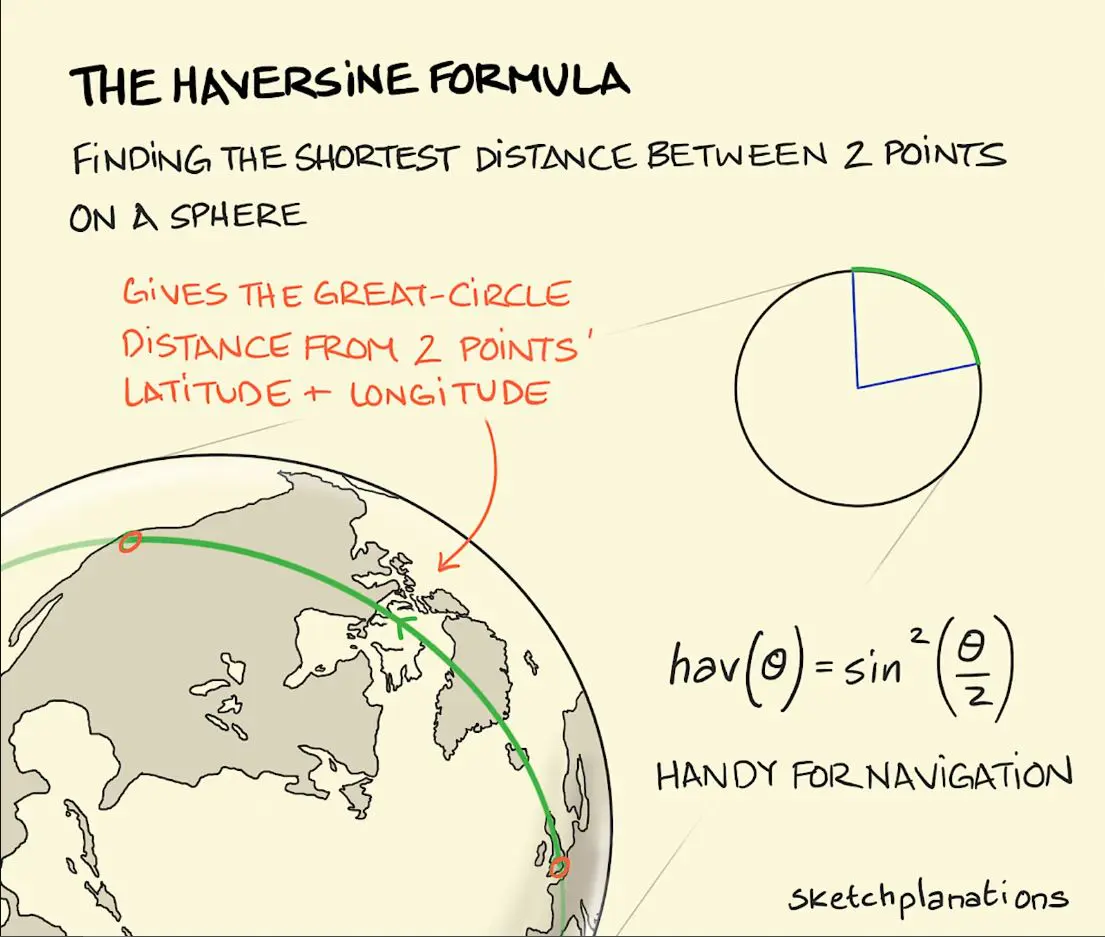
Here is simple numpy implementation:
def haversine_np(
lat_long_deg_1: npt.NDArray,
lat_long_deg_2: npt.NDArray,
radius: float = RADIUS_EARTH_KM,
) -> npt.NDArray:
"""
Calculate the great circle distance between two points on a sphere
ie: Shortest distance between two points on the surface of a sphere
"""
lat_1, lon_1, lat_2, lon_2 = map(
np.deg2rad,
[
lat_long_deg_1[:, 0],
lat_long_deg_1[:, 1],
lat_long_deg_2[:, 0],
lat_long_deg_2[:, 1],
],
)
d = (
np.sin((lat_2 - lat_1) / 2) ** 2
+ np.cos(lat_1) * np.cos(lat_2) * np.sin((lon_2 - lon_1) / 2) ** 2
)
arc_len = 2 * radius * np.arcsin(np.sqrt(d))
return arc_len
NOTE: this post is framed around an example of haversine distance, but this technique applies to many different scenarios. Maybe you need to measure the levenshtein edit distance between strings or a compatibility score between potential wedding guests :)
I was shocked to discover that most clustering tool kits (scikit-learn) do NOT support custom distance metrics. Those that do (NLTK) are extremely slow, because they execute a provided distance function for every pair of points individually. This is ok for clustering a handful of data points…. but will NOT scale to millions of samples. Instead I wrote a pseudo vectorized implementation like so:
Nearest-Neighbor
For my problem, I needed relatively even spacing between clusters. For this a nearest-neighbor algorithm was a better choice. While not completely vectorized, this approach scales more linearly than exponentially.
def haversine_cluster(
points_lat_long_deg: npt.NDArray,
centroids_lat_long_deg: npt.NDArray,
trace: bool = False,
) -> npt.NDArray:
"""Cluster points to the closest centroid based on haversine dist
Args:
points_lat_long_deg (npt.NDArray): the data points to cluster, shape (n, 2)
centroids_lat_long_deg (npt.NDArray): the cluster centroids, shape (k, 2)
trace (bool, optional): If True, display progress bar. Defaults to True.
Returns:
(npt.NDArray): labels (cluster indices) for each data point
"""
# Cluster the data points to the nearest "cluster" based on haversine dist
n = points_lat_long_deg.shape[0]
k = centroids_lat_long_deg.shape[0]
# Assign centroids based on minimum haversine distance
diff = np.zeros((n, k))
for i in tqdm(range(k), disable=not trace):
diff[:, i] = haversine_np(
points_lat_long_deg, centroids_lat_long_deg[np.newaxis, i, :]
)
labels = diff.argmin(axis=1) # n,
return labels
# Pre-define grid of clusters
# TODO: this could be further improved by initializing equidistant centroids (http://extremelearning.com.au/evenly-distributing-points-on-a-sphere/)
# For now, just prune unpopulated clusters afterwards
n_div_lat = 250
n_div_long = 500
cluster_div_lat = np.linspace(np.min(data[:,0]), np.max(data[:,0]), n_div_lat)
cluster_div_long = np.linspace(np.min(data[:,1]), np.max(data[:,1]), n_div_long)
centroids = np.zeros((cluster_div_lat.shape[0]*cluster_div_long.shape[0], 2))
for i, lat in enumerate(cluster_div_lat):
for j, long in enumerate(cluster_div_long):
centroids[i * n_div_long + j] = [lat, long]
# Then cluster the data points to the nearest "cluster" based on haversine dist
labels = haversine_cluster(
# use only a subset of data points... since this is expensive
points_lat_long_deg=data,
centroids_lat_long_deg=centroids,
trace=True
)
# Remove any clusters that have no data points...
# this reduces the final number of clusters while keeping an even spacing
populated_centroid_idxs = np.array(sorted(np.unique(labels)))
centroids = centroids[populated_centroid_idxs, :]
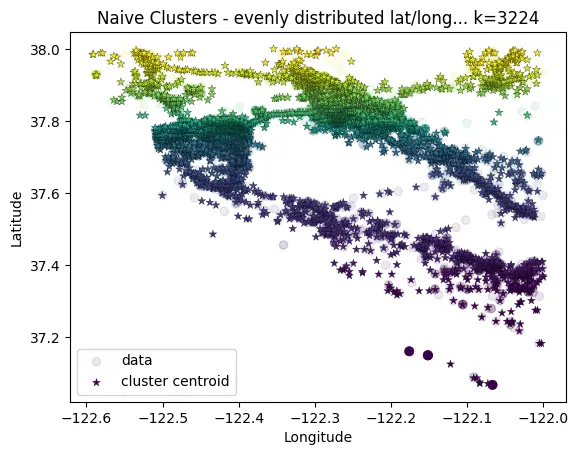
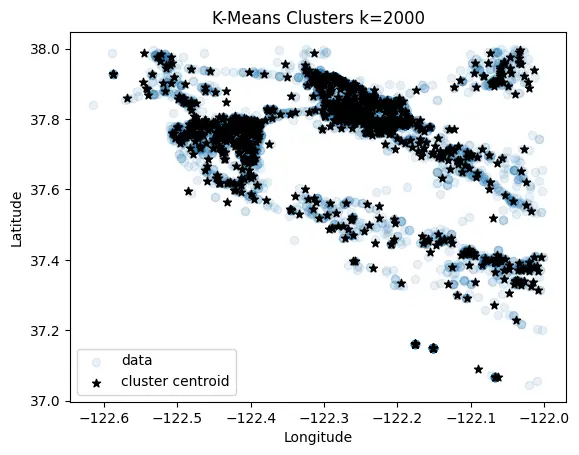
K-Means
A k-means implementation just for grins :)
def k_means(data, k:int=50, num_iter:int=50, seed:int=42, trace: bool=False):
np.random.seed(seed)
n, d = data.shape
data_lla = np.stack([data[:,0], data[:,1], np.zeros((n))]) # 3, n
# Initialize centroids as random selection of data points
centroids = data[np.random.choice(n, k, replace=False)] # k, d
diff = np.zeros((n,k))
for _ in tqdm(range(num_iter), disable=not trace):
# Assign centroids based on minimum haversine distance
for i in range(k):
diff[:, i] = haversine_np(data, centroids[np.newaxis, i,:])
labels = diff.argmin(axis=1) # n,
# Update the centroids to be the projected centroid of the members of each cluster
for i in range(k):
member_idxs = np.argwhere(labels==i).squeeze()
members = data_lla[:, member_idxs] # 3, x
if members.shape[-1] == 0:
# empty cluster... don't update
continue
centroid = calculate_centroid_geo(lat_long_alt=members.swapaxes(0,1)) if members.ndim > 1 else members
centroids[i] = centroid[:2]
break
return centroids
For more details see associated repository https://github.com/dcyoung/ml-triangulation

Comments